Simple simulation of birth and death
Introduction
Our goal is to simulate the birth and death of three cell species (A, B, C). The frame bellow is a result of this simulation: three different cells processes (light grey, grey and black) are being simulated.
Empirical data
For each cell we know empirically that:
- Every 2 seconds a new cell of type A is born.
- Every 15 seconds a cell of type A dies.
- Every 5 seconds a new cell of type B is born.
- Every 10 seconds a cell of type B dies.
- Every 4 seconds a new cell of type C is born.
- Every 30 seconds a cell of type C dies.
Assumptions
We consider that the birth of a cell is an event that occurs continuously and independently at a constant average rate. Continuously because we do not consider things like temperature and independently because the birth of one cell does not affect the birth of another cell.
A tool: Poisson process
This birth and death processes of our cells are called stochastic processes.They can be modelised with a Poisson process.
Our assumptions defined previously are in fact conditions to use a Poisson process:
- Events occurs continuously.
- Events occurs independently.
- Events occurs at a constant average rate (homogeneous Poisson process).
In a Poisson process the time between two events is described with an exponential distribution of parameter lambda (λ). It is called the rate parameter and it is equal to 1 devided by the desired mean value.
So in the case of our cells λ can be valued as follows:
- Every 2 seconds a new cell of type A is born: λ = 1/2.
- Every 15 seconds a cell of type A dies: λ = 1/15.
- Every 5 seconds a new cell of type B is born: λ = 1/5.
- Every 10 seconds a cell of type B dies: λ = 1/10.
- Every 4 seconds a new cell of type C is born: λ = 1/4.
- Every 30 seconds a cell of type C dies: λ = 1/30.
The exponential distribution
We said that our process is using an exponential distribution of parameter λ to describe the time between each pair of birth and each pair of death.
A probability distribution assign a probability to each outcomes of a random experiment. The outcome of an experiment is a random variable called x. We can define for a probability distribution a function F(x) that will output a probability for a given value of x. This function F is called a cumulative density function.
Cumulative density function
The exponential distribution is characterized by its density function which is:
This cumulative density function is the sum (i.e. the integration) of the probability density function. It describes the probability that the random variable x will takes a values less or equal to a given X.
This is a plot of the cumulative distribution function of the exponential distribution for λ = 1/30.
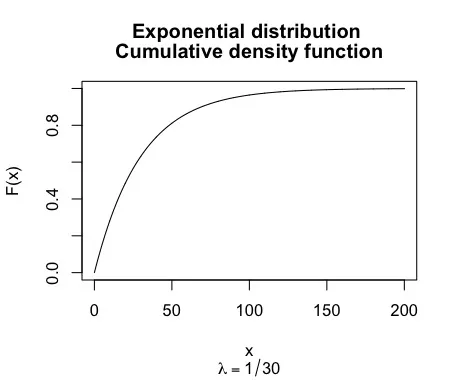
If we consider that the y-axis is the time we can intuitively see that the more we wait the more the probability of the outcome is higher.
Reading the cumulative density function in reverse
Let’s say that the y-axis describes the time in seconds. The x-axis is valued between 0 and 1.
So each time we want to know how much time we have to wait before seeing a positive outcome for the experiment we can randomly choose a number between 0 and 1 that will be the value of F(x) and read the value of the corresponding x. In the end value of x will give us the time we have to wait before seeing an occurrence of the event.
We can use the cumulative density function equation to express x knowing the value of F(x):
With
Testing
In order to test this we are going to generate a grand number of values for λ = 1/30. That means that the events on the Poisson process occurs at a constant average rate of 30 seconds. If we calculate the mean of the generated numbers we should get a value likely equal to 30.
To do so we are going to use Python which provide a built-in method in the random module which return exponentialy distributed number for a given λ parameter.
>>> import random
>>> lambda_parameter = 1/30.0
>>> random.expovariate(lambda_parameter)
20.401058019178667
>>> values = [random.expovariate(lambda_parameter) for i in xrange(100000)]
>>> mean = sum(values) / len(values)
>>> mean
>>> 29.821100014718574The mean is likely equal to 30! Our simulated process has events that occurs at the right mean rate.
Building a cell simulation with Javascript
Javascript does not provide a expovariate function. Here is an implementation for it:
Random = {};
Random.expovariate = function (lambda) {
return -Math.log(1 - Math.random()) / lambda;
};Given the born and death rate of the cell we can determine with the expovariate function a time: the time to the next birth of a cell and the time to live of this cell. In Javascript the setTimout function can execute a function after a specified amount of time. We can use it to call a birth function and a death function with the given calculated times. SetTimeout use milliseconds: we need to multiply by 1000 our rate values.
var makeCellProcess = function (cellSymbol, bornRate, deathRate) {
var start = function () {
return function () {
var timeToNextBirth = Random.expovariate(1 / bornRate),
timeToLive = Random.expovariate(1 / deathRate),
cell = Grid.placeCell(cellSymbol);
setTimeout(end(cell), timeToLive);
setTimeout(start(), timeToNextBirth);
};
};
var end = function (cell) {
return function () {
cell.remove();
};
};
// Calling start to initialize the process.
start()();
};
makeCellProcess(cellASymbol, 1000, 15000);Conclusion
- Have a look to the annotated source of the cell simulation.
- You can simply simulate events occurring with known mean interval with a Poison process.
- A probability distribution gives the probability of a random variable x for a given experiment.
- A cumulative distribution function output the probability that a random variable X will be found at a value less than or equal to x.